Large nanopores, realizable by standard lithographic techniques, can be bio-functionalized to obtain a final device with dimensions compatible with single molecule sensing applications, and selective for different kind of interactions between probe and target molecules.
http://nanotechweb.org/cws/article/lab/42519
Single molecule sensing with nanopores is a rapidly developing field which exerts a special fascination on the scientific community, both for the potential revolutionary effect it can have on bioanalytics and diagnostics, and for its striking intrinsic interdisciplinary. Starting from the first papers mainly devoted to illustrate the properties of protein pores, much work has been done to realize engineered devices based on solid state nanopores, with specific characteristics and sensing abilities. Nowadays the spectrum of proposed nanopore applications is quite large, most of them being based on electrophoretic current measurements during molecules translocation, i.e. on a simple functioning principle which does not need molecular labeling and optical detection. Nanopore sensing can thus provide a low cost, fast processing and high throughput alternative to current DNA analysis and sequencing techniques.
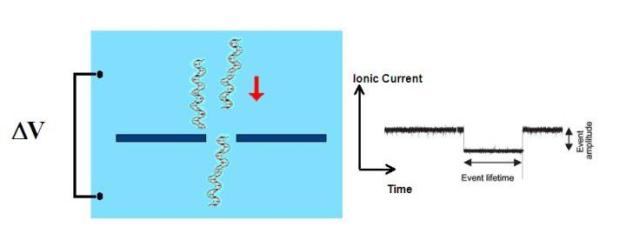
Figure
1. "Coulter counter" principle of functioning of nanopore
devices
The specific sensing mechanism merges coulter counting and single current recording (figure 1).: the nanopore conductance is modulated during the passage of single molecules by steric or electrostatic effects. Nevertheless, nanopores properties can be finely tuned by introducing artificial binding and recognition sites or, generally speaking, by chemically modifying their surface properties (figure 2).
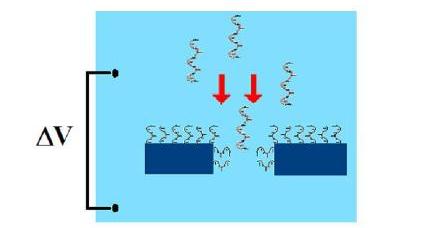
Figure 2. Chemically modified nanopores for selective sensing
We realized
a new class of biosensor devices (figure 3) based on bio-functionalized
SiN nanopores (figure 4) able to detect different kind of interactions
between probe molecules, chemically attached to the pore, and target
molecules*. The great potentiality of this approach resides in the fact
that the bio-functionalization of a quite large pore (up to 30-60 nm),
allows a sufficient diameter reduction for the attainment of single
molecule sensing dimension and the selective activation, without the
need for further material deposition, such as metal or oxides, or localized
surface modification.
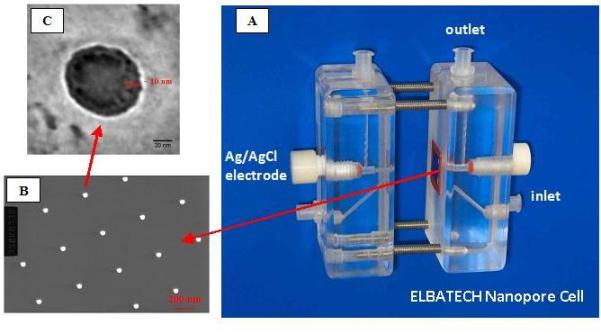
Figure
3.NPA
device based on an array of bio-functionalized nanopores for single
molecules analysis. A: Measring cell realized in plexyglass and containing
the central housing for the Si/SiN chip with the FIB drilled nanopores,
the microfluidic system, two reservoirs for the ionic solution and a
couple of Ag/AgCl electrodes to apply the voltage and collect the nano-ampere
current; B: The surface of the SiN thin membrane (20 nm thick, 80x80
mm large) with the array of 60 nm large FIB fabricated nanopores; C:
A single nanopore chemically funcionalized with probe molecules for
the selective recognition of target molecules.
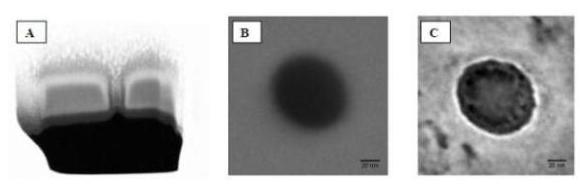
Figure
4.
Cross section of a nanopore fabricated by FIB milling on a silicon nitride
100 nm thick membrane, A SEM image of the nanopore before, B,
and after, C, the functionalization with 45-mer oligonucleotides
[4,5]
The single molecule sensing ability of our device was recently demonstrated by registering translocation events of -DNA molecules through a 40 nm diameter FIB drilled nanopore bio-functionalized with 45-mer oligonucleotides (figure 5).
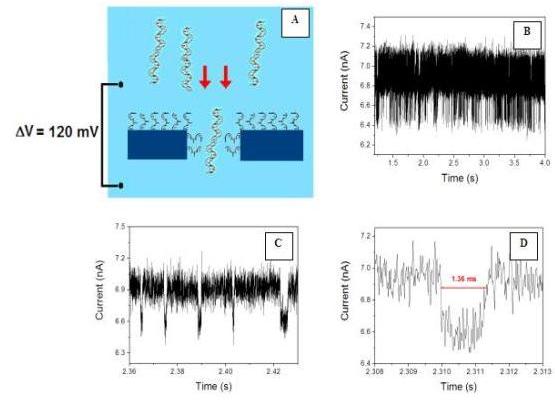
Figure
5. A: Scheme of the translocation experiment: l-DNA target molecules
are electroforetically drawn through a nanopore functionalized with
45-mer oligonucleotides; B: Current trace registered during the
passage of the molecules: C. D: progressive magnification of
the trace to visualize a single event.
Our
results demonstrate that it will be possible, in a near future, to conceive
and design devices for parallel analysis of biological samples, made
of arrays of nanopores, fabricated by standard lithographic techniques
and finely tuned and resized by bio-functionalization.
This kind of hybrid biosensors, which could be integrated into more
complex microfluidic devices and lab-on-a chips, promise to have important
applications in the field of molecular diagnosis.
We made a step further in this direction with the Polimeric nanochannel device, which is based on chemically modified nanochannels created on a polymeric substrate. The possibility to realize a polymeric nanopore offers many advantages over solid state technologies such as really cheap, rapid and easy fabrication and integrability. Combining nanofabrication and soft lithography it is possible to create a nanochannel device as shown in figure 6 which can be functionalized with oligonucleotides by means of a specifically developed protocol (see figure 7).
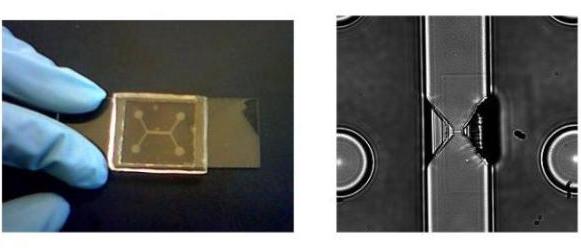
Figure6.
On the left, a picture of the final polymeric device; on the right,
an image of a nanochannel linking the inlet and the outlet of the device
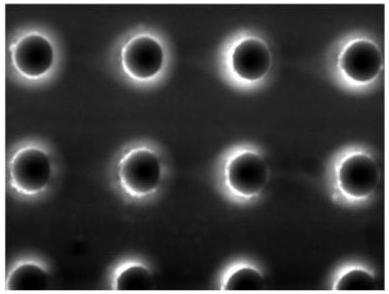
Figure 7. Fluorescence image of the microchannels along the inlet and the outlet, where cylindrical pillar are presents, showing the presence of Fitc-labelled DNA
"WHO'S WHO" OF THE PROJECT
Valentina Mussi is a staff researcher,
responsible of the nanopore project. Dr. Luca
Repetto and Giuseppe Firpo realized
the FIB fabrication and SEM imaging activity. The team is directed by
Professor Ugo Valbusa of the Physics Department
of the University of Genova. Dr. Paola Fanzio
and Chiara Manneschi are Ph.D. students
of the University of Genova. The project directly involves Dr. Paola
Scaruffi, Dr. Sara Stigliani and
Prof. Gian Paolo Tonini of the National
Institute of Cancer Research of Genova.
Publications related to the project:
[1] V. Mussi, P. Fanzio, L. Repetto, G. Firpo, U. Valbusa, P. Scaruffi,
S. Stigliani, G. Tonini "Solid State nanopores for gene expression
profiling", Superlattices and Microstructures 46 (2009)
59-63.
[2] V. Mussi, P. Fanzio, L. Repetto, G. Firpo, S. Stigliani, P. Scaruffi, G.P. Tonini and U. Valbusa, "DNA-functionalized nanopores for single molecule analysis", Nanotecnology, 2009, Proceedings of IEEE-NANO 2009, 9th IEEE Conference, Publication Year: 2009, Pages 682 - 684.
[3] P. Fanzio, E. Angeli, V. Mussi, U. Valbusa, P. Rivolo, F. Frascella, F. Pirri, "Development of a polymeric device for gene expression profiling", Nanotecnology, 2009, Proceedings of IEEE-NANO 2009, 9th IEEE Conference, Publication Year: 2009, Pages: 613 - 615.
[4] V. Mussi, P. Fanzio, L. Repetto, G. Firpo, P. Scaruffi, S. Stigliani, GP. Tonini, U. Valbusa, "DNA-functionalized solid state nanopore for biosensing", Nanotechnology 21 (2010) 145102.
[5] V. Mussi, P. Fanzio, L. Repetto, G. Firpo, P. Scaruffi, S. Stigliani, M. Menotta, M. Magnani, G. P. Tonini and U. Valbusa, "Electrical characterization of DNA-functionalized solid state nanopores for bio-sensing", submitted to Journal of Physics: Condensed Matter.
*Italian Patent # RM2009A000450, 4th September 2009, V. Mussi. P. Fanzio, U. Valbusa, L. Repetto, G. Firpo, S. Stigliani, P. Scaruffi, G.P. Tonini, V. Mussi. P. Fanzio, U. Valbusa, L. Repetto, G. Firpo, S. Stigliani, P. Scaruffi, G.P. Tonini, "Chip nanoforato di nitruro di silicio per l'analisi di profili di espressione genica e relativi biosensori" ("Silicon nitride nanopore chip for gene expression profiling and biosensing");
How to reach us
Last update
6/23/10
