Genotype-Haplotype Microscopy (GHM).
GHM project can be divided in three different research branches:
Combined determination of genotype and haplotype by using probes which are detectable on single DNA molecule at a distance as close as 10 nucleotides.
The chromosomes in human cells occur in pairs (with the exception
of the sex cells). One member of each chromosome pair is inherited
from a person's father; the other member of the pair is inherited
from that person's mother. But chromosomes do not pass from each generation
to the next one as identical copies. Rather, the chromosome pairs
undergo a process known as recombination. The members of each chromosome
pair come together and exchange pieces. The result is a hybrid chromosome
containing pieces from both members of a chromosome pair, and this
hybrid chromosome is passed on to the next generation. Over the course
of many generations, segments of the ancestral chromosomes are through
repeated recombination events. Some of the segments of the ancestral
chromosomes occur as regions of DNA sequences that are shared by multiple
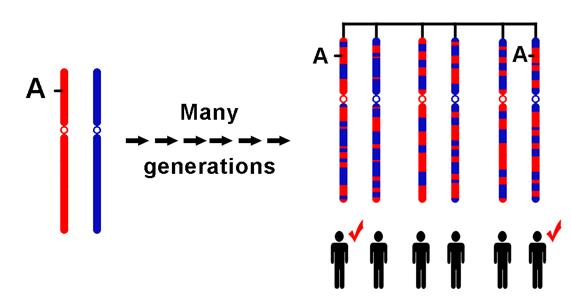
individuals (Figure 1).
Figure
1. The two ancestral chromosomes being scrambled through recombination
over many generations to yield different descendant chromosomes. If
a genetic variant marked by the A on the ancestral chromosome increases
the risk of a particular disease, the two individuals in the current
generation who inherit that part of the ancestral chromosome will
be at increased risk. Adjacent to the variant marked by the A are
many SNPs that can be used to identify the location of the variant.
These
segments are regions of chromosomes that have not been broken up by
recombination, and they are separated by places where recombination
has occurred. These segments are the haplotypes that enable geneticists
to search for genes involved in diseases and other medically important
traits.
The most common form of variation in DNA sequence is the single-nucleotide
polymorphism (SNP); SNPs are present at a frequency of about 1 in 1,000
nucleotides in humans.
We are developing a method to haplotyping by direct visualization of
polymorphic sites on DNA molecules. Our approach utilizes atomic force
microscopy to detect directly the polymorphic sites in DNA fragments.
In principle, the methods of DNA sequencing give us information on the
presence of SNPs along the DNA strand, that can be used to assemble
all the combinations of SNPs that identify the various haplotypes, choosing
an appropriate DNA fragment as standard and hybridizing it with DNA
under investigation, we obtain DNA-heteroduplex strands (Figure 2).
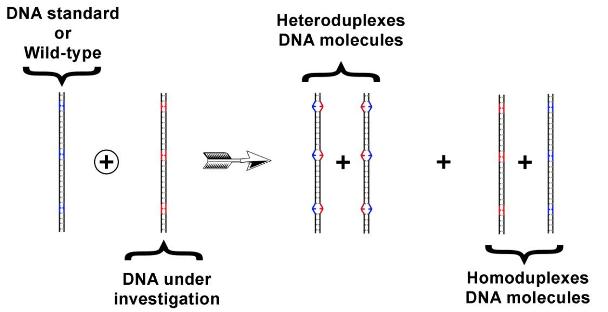
Figure 2. Heteroduplexes-DNA formation. The heteroduplexes are characterized by the presence of one o more mismatches, due to the presence of different nucleotides. We use a protein mismatch repairing to mark the mismatch present in such DNA-heteroduplex molecules.
We are
evaluating various approaches to detect the mismatch position along
DNA stands. Firstly, we have investigated heteroduplexes of DNA with
only one mismatch. Presently we are investigating heteroduplexes DNA
molecules with two mismatches.
Construction of a genotype-haplotype microscope (GHM).
Simultaneously, to the haplotype determination, we are developing a
scanning probe microscope (Figure3), based on AFM technology, which
will be dedicated to investigate the DNA haplotype, the protein-protein
interactions and the interactions between DNA and proteins.
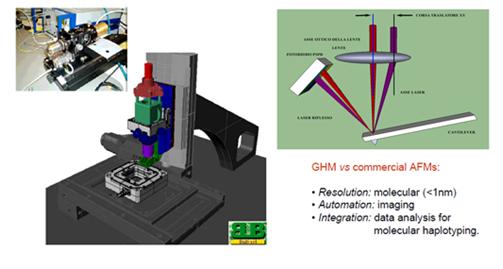
Figure
3.CAD
project of the GHM (middle), head of the microscope (left) and a scheme
of the optical beam deflection technique used in GHM setup (right)..
We are building a prototype of the GHM using Physik Instrumente (PI) components (controller, translators, actuators and stages). Tests of cantilever thermal noise have given better results than those characteristic of commercial AFM (Figure 4).
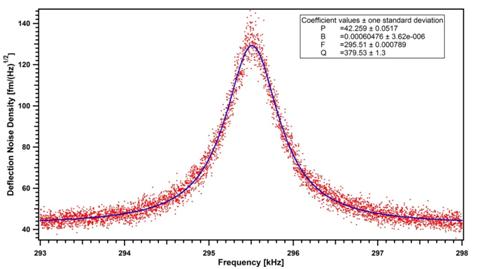
Figure
4.
Frequency spectra of cantilever Brownian motion measured in air.
With this prototype we have acquired various force-distance curves on Si surfaces, which are perfectly reproducible and show the well known regions of the force-distance curves (Figure 5).
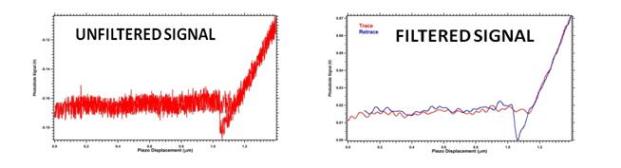
Figure 5. Force-distance curves recorded by GHM prototype: unfiltered signal (left) and filtered signal (right).
Study of protein-protein and dna-protein interaction with GHM microscope.
The design
and realization of protein nano-arrays aim to develop devices able to
detect protein markers in very small and dilute samples, in which the
protein markers concentration reaches values of the order of 10-16 M.
Scanning probe microscopy is a powerful tool in studies that involve
biological systems, our aim is to use GHM microscope to study the ligand-receptor
interaction. The critical point with atomic force microscopy techniques
is the roughness of the substrate surface: if the roughness of the surface
is too high, it is impossible to recognize the protein bound to the
surface. Therefore, we have looked for an appropriate method to functionalize
the substrate surface. The first step is a formation of a layer of 3-aminopropyltriethoxysilane
(APTES) on freshly cleaved mica surface. Figure 6 shows an AFM image
of the mica-APTES surface with a low roughness.
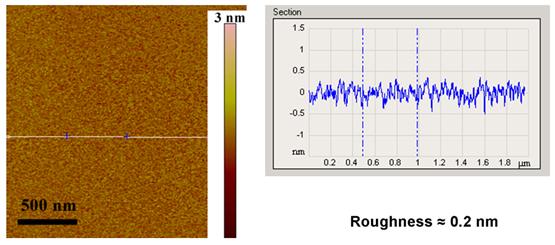
Figure 6: AFM Tapping mode image of mica-APTES surface, that shows a very low roughness
After the APTES functionalization we have added another layer of a linker substance* able to bind properly Fc portion of the antibody and to leave the antigen binding sites in the antibody Fab portion accessible (Figure 7).
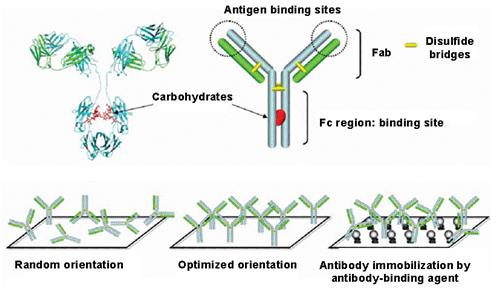
Figure 7: Scheme of the antibody immobilization on the mica-APTES-linker
surface.
The roughness of the mica-APTES-linker surface is nearly unchanged, so that it is possible to study the antibody-antigen interaction by scanning probe microscopy.
"WHO'S WHO" OF THE PROJECT
Vincenzo
Ierardi
is a staff researcher, responsible of the GHM project. The team is directed
by Professor Ugo Valbusa of the Physics Department
of the University of Genova.
The project directly involves:
" Dr. Francesca Giacopelli and Prof.
Roberto Ravazzolo of the Gaslini Institute
of Genova for the studies on the DNA haplotype,
" Dr. Paolo Domenichini of the BiuBi
s.r.l. Genova for the realization of GHM microscope,
" Dr. Francesca Ferrera and Prof. Gilberto
Filaci of the CEBR-Center of Excellence for Biomedical Research
of the University of Genova for the studies on ligand-receptor complexes.
* F. Ferrera, V. Ierardi, E. Millo, G. Filaci, G. Damonte, F. Indiveri,
U Valbusa, "Development of a new procedure for controlled antibodies
immobilization useful for the construction of an antibody nanoarray allowing
the measurement of antigen-antibody immune-complex via label free technique",
Italian Patent, 17/11/2009;
How to reach us
Last update
6/23/10
